DAP-seq(DNA亲和纯化测序)
260+物种,4000+转录因子实战经验,无需抗体
高通量检测转录因子或DNA结合蛋白在基因组上的结合位点
助力客户发表高分文章Cell,Science,Molecular Plant,Plant Biotechnology Journal,Journal of Advanced Research,Plant Cell,PNAS,Plant Communications,Journal of Integrative Plant Biology,Molecular Horticulture,New Phytologist,International Journal of Biological Macromolecules,Horticulture Research,Current Biology,Plant Physiology等。
- 技术简介
- 服务列表
- 服务内容
- 经验分享
- 常见问题
- 客户文章
在功能基因组学和表观遗传学研究中,转录因子结合位点(TFBS)的发掘一直是研究热点。传统的ChIP-seq(染色质免疫共沉淀测序)方法,在抗体质量很好的情况下能够有效检测到TFBS。然而,好的抗体可遇不可求,这限制了ChIP-seq更广泛的应用。
DAP-seq技术的出现,使TFBS 的研究不再局限于物种,不再受抗体质量的限制,为生命科学领域转录因子的研究提供了新的有效工具。
DAP-seq与ChIP-seq技术对比
| 技术名称 | DAP-seq | ChIP-seq |
| 实验模式 | 体外 | 体内 |
| 是否需要特异性抗体 | 否 | 是 |
| 是否适用于非模式物种 | 是 | 否 |
| 时间成本 | 低 | 高 |
| 是否高通量 | 是 | 否 |
蓝景科信拥有260+物种,4000+转录因子的实验经验,周期短,口碑好,助力客户发表高分文章Cell,Science,Molecular Plant,Plant Biotechnology Journal,Journal of Advanced Research,Plant Cell,PNAS,Plant Communications,Journal of Integrative Plant Biology,Molecular Horticulture,New Phytologist,International Journal of Biological Macromolecules,Horticulture Research,Current Biology,Plant Physiology等。
参考文献:
O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell. 2016. 165(5):1280-1292. doi: 10.1016/j.cell.2016.04.038.
![]() 2016-Cell-DAP Seq-Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape.pdf
2016-Cell-DAP Seq-Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape.pdf
Bartlett A, O'Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017. (8):1659-1672. doi: 10.1038/nprot.2017.055.
![]() 2017-Nature Protocols-Mapping genome-wide transcription-factor binding sites using DAP-seq.pdf
2017-Nature Protocols-Mapping genome-wide transcription-factor binding sites using DAP-seq.pdf
| 服务项目 | 周期 | 交付结果 | 报价 |
| 蛋白表达载体构建 | 1-2周 | 构建载体的测序结果 实验过程图 原始测序数据 分析结果 | 详细报价请电询400-6187099 或15632249798 |
| 蛋白无细胞表达 | 1-2周 | ||
| DAP-seq文库构建 | 1周 | ||
| DNA亲和纯化 | 1-2周 | ||
| 上机测序 | 2周 | ||
| 标准数据分析 | 2周 |

| 实验流程 |

| 生信分析 |
|
|
| 项目可行性分析 |
开展项目之前,我们会根据您具体的转录因子做可行性分析报告,供您参考,从多个方面进行可行性分析,包括转录因子分子量,亚细胞定位预测,跨膜区预测,蛋白质结构域预测、翻译后修饰预测,并且根据文献报道和我们的经验来进行可行性分析。

| 已做物种 | ||||||||
| 植物 | ||||||||
| 粮食和经济作物 | ||||||||
| 大豆 | 大麦 | 谷子 | 高粱 | 玉米 | 水稻 | 小麦 | 燕麦 | 苦荞 |
| 花生 | 芝麻 | 油菜 | 甘蓝型油菜 | 藜麦 | 菜豆 | 豌豆 | 木薯 | 马铃薯 |
| 甘薯 | 红薯 | 茶树 | 棉花 | 橡胶树 | 烟草 | 麻疯树 | 油桐 | 甜菜 |
| 圆果种黄麻 | 毛竹 | 麻竹 | 桑树 | 甘蔗 | 博落回 | 花椒 | 橡胶草 | 翅果油树 |
| 大麻 | 可可 | 文冠果 | ||||||
| 蔬菜 | ||||||||
| 菠菜 | 番茄 | 黄瓜 | 茄子 | 胡萝卜 | 冬瓜 | 生菜 | 芥蓝 | 茎瘤芥 |
| 小白菜 | 不结球白菜 | 菜心 | 甘蓝 | 西葫芦 | 香椿 | 莲 | 辣椒 | 龙须菜 |
| 蔓菁 | 白菜 | 白菜型油菜 | 青梗菜 | |||||
| 水果 | ||||||||
| 菠萝 | 柑橘 | 荔枝 | 苹果 | 葡萄 | 草莓 | 猕猴桃 | 香蕉 | 橙子 |
| 柿子 | 杏 | 桃 | 樱桃 | 西瓜 | 甜瓜 | 无花果 | 芒果 | 板栗 |
| 核桃 | 冬枣 | 枣 | 山金柑(金柑) | 毛葡萄 | 软枣猕猴桃 | 椪柑 | 杨梅 | 凤梨 |
| 椰枣 | 火龙果 | 梨 | 李 | 龙眼 | 石榴 | 毛酸浆 | ||
| 花卉和观赏植物 | ||||||||
| 菊花 | 玫瑰 | 牡丹 | 月季 | 蓝花耧斗菜 | 夏堇 | 紫薇 | 芍药 | 小兰屿蝴蝶兰 |
| 百合 | 甘野菊 | 百岁兰 | 建兰 | 向日葵 | 甘菊 | 白木香 | 梅 | 大马士革黑种草 |
| 二色补血草 | 海棠 | 珙桐 | ||||||
| 药用植物 | ||||||||
| 丹参 | 黄连 | 青蒿 | 人参 | 短小蛇根草 | 黄花蒿 | 铁皮石斛 | 枸杞 | 大叶秦艽 |
| 大叶秦艽 | 灰毡毛忍冬 | 枳 | 藏红花 | 金银花 | 粉葛 | 香橼(香圆) | 党参 | 杜仲 |
| 广藿香 | 黄芩 | 雷公藤 | 三叶青 | 五味子 | ||||
| 林木 | ||||||||
| 大青杨 | 旱柳 | 落叶松 | 马尾松 | 毛白杨 | 毛果杨 | 闽楠 | 青钱柳 | 小垫柳 |
| 楸树 | 栓皮栎 | 油松 | 小黑杨 | 小叶杨 | 银杏 | 光皮桦 | 胡杨 | 灰杨 |
| 欧美杨 | 欧洲云杉 | 杉木 | 木荷 | 山桃 | 杂交枫香 | 滇杨 | 山新杨 | 717 杨 |
| 刚毛柽柳 | 84K杨 | 桉树 | 白桦 | 杜梨 | 酸枣 | 无患子 | ||
| 草类和牧草 | ||||||||
| 稗草 | 狗尾草 | 高加索三叶草 | 结缕草 | 鸭茅 | 黑麦草 | 柳枝稷 | 千金子 | 短芒大草 |
| 蒺藜苜蓿 | 紫花苜蓿 | 杂花苜蓿 | 百脉根 | 星星草 | ||||
| 藻类 | ||||||||
| 紫菜 | 球等鞭金藻 | 三角褐指藻 | 中带鼓藻 | 圆柱拟脆杆藻 | 浒苔 | |||
| 其他植物 | ||||||||
| 拟南芥 | 盐芥 | 疏花水柏枝 | 伴矿景天 | 黄花棘豆 | 小立碗藓 | 苔藓 | 地钱 | 角果碱蓬 |
| 动物 | ||||||||
| 小鼠 | 驴 | 羊 | 梅花鹿 | 斑点叉尾鮰 | 团头鲂 | 飞蝗 | 烟粉虱 | 草地贪夜蛾 |
| 褐飞虱 | 斜纹夜蛾 | 二化螟 | 蜜蜂 | 华贵栉孔扇贝 | 曼氏血吸虫 | 新孢子虫 | ||
| 真菌 | ||||||||
| 糙皮侧耳 | 草菇 | 灰盖鬼伞 | 元蘑 | 金针菇 | 高卢蜜环菌 | 猪苓真菌 | 灵芝 | 虫草 |
| 蝗绿僵菌 | 大丽轮枝菌 | 拟轮枝镰孢菌 | 亚洲镰刀菌 | 禾谷镰刀菌 | 意大利青霉 | 草酸青霉 | 里氏木霉 | 金黄壳囊孢 |
| 灰霉菌 | 裂殖壶菌 | 疫霉 | ||||||
| 细菌 | ||||||||
| 生氮假单胞菌 | 巴西固氮螺菌 | 根瘤菌 | 类球红细菌 | 红杆菌科细菌 | 伯克霍尔德菌 | 大肠杆菌 | 路德维希肠杆菌 | 肺炎克雷伯菌 |
| 成团泛菌 | 沙门氏菌 | 铜绿假单胞菌 | 美人鱼发光杆菌 | 杀鱼爱德华氏菌 | 嗜水气单胞菌 | 布鲁氏菌 | 脓肿分枝杆菌 | 结核分枝杆菌 |
| 嗜热厌氧杆菌 | 解淀粉芽孢杆菌 | 地衣芽胞杆菌 | 水稻白叶枯病菌 | 集胞藻 | ||||
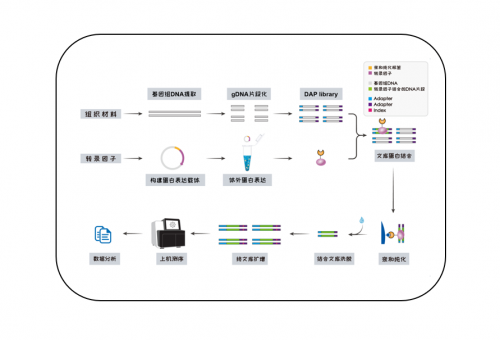
1.DAP-seq原理是什么,技术流程是什么,能帮我解决什么样的问题?
原理:体外表达的蛋白和DNA进行亲和纯化,将与蛋白结合的DNA洗脱后进行高通量测序。
技术流程:将编码转录因子的CDS序列构建到含有亲和标签的载体中,构建蛋白表达载体,进行体外蛋白表达,形成转录因子和亲和标签的融合蛋白;提取样品的基因组DNA,构建DNA文库,然后将体外表达的带有亲和标签的转录因子和DNA文库进行结合,随后把结合的DNA洗脱后上机测序。
能帮助您快速找到转录因子的结合位点,寻找转录因子调控的靶基因。
技术服务流程:

2.需要提供什么材料?
需要您提供
(1)组织材料或者是提取好的基因组DNA;
(2)含有转录因子CDS序列的质粒。
3.分析结果包括哪些内容?
蓝景科信DAP-seq的生信分析包括以下内容:
1. 对原始数据进行去除接头、污染序列及低质量 reads 的处理
2. 数据产出统计
3. 参考序列比对分析
4. 测序reads富集区域扫描(peak calling)
5. Peak在基因功能元件上的分布统计
6. Peak序列模式发掘(motif search)
7. 已知motif注释
8. Peak相关基因鉴定
9. Peak相关基因的GO和KEGG富集分析
10. 测序数据的可视化分析
4.实验的成功率怎么样?
不同转录因子家族的成功率不同,请参考不同转录因子家族的DAP-seq成功率:
不同转录因子家族成功率 | |||
转录因子 家族类型 | 该家族已做 转录因子的数量 | 成功鉴定到Motif的 转录因子数量 | 该家族转录因子的 成功率 |
C2H2 | 151 | 27 | 18% |
bHLH | 137 | 18 | 13% |
AP2-EREBP | 133 | 74 | 56% |
C3H | 129 | 9 | 7% |
MYB | 116 | 55 | 47% |
MADS | 86 | 10 | 12% |
NAC | 76 | 51 | 67% |
MYB-related | 71 | 26 | 37% |
WRKY | 65 | 34 | 52% |
ND | 60 | 5 | 8% |
Homeobox | 43 | 13 | 30% |
ABI3-VP1 | 40 | 7 | 18% |
bZIP | 38 | 29 | 76% |
G2-like | 37 | 17 | 46% |
LOB-AS2 | 35 | 8 | 23% |
Orphan | 35 | 3 | 9% |
C2C2-CO-like | 34 | 2 | 6% |
C2C2-DOF | 32 | 21 | 66% |
C2C2-GATA | 28 | 13 | 46% |
HB | 27 | 10 | 37% |
Trihelix | 27 | 13 | 48% |
TCP | 26 | 13 | 50% |
mTERF | 23 | 1 | 4% |
GeBP | 19 | 2 | 11% |
HSF | 17 | 10 | 59% |
SBP | 16 | 8 | 50% |
ZF-HD | 14 | 6 | 43% |
ARF | 12 | 3 | 25% |
CCAAT-HAP5 | 12 | 2 | 17% |
FAR1 | 12 | 1 | 8% |
FHA | 12 | 1 | 8% |
HMG | 12 | 1 | 8% |
CCAAT-HAP3 | 11 | 2 | 18% |
PLATZ | 11 | 1 | 9% |
ARID | 10 | 5 | 50% |
LIM | 10 | 1 | 10% |
BSD | 9 | 1 | 11% |
CPP | 8 | 4 | 50% |
GRF | 8 | 2 | 25% |
REM(B3) | 8 | 1 | 13% |
SRS | 8 | 1 | 13% |
BBR/BPC | 7 | 3 | 43% |
E2F-DP | 7 | 4 | 57% |
BZR | 6 | 4 | 67% |
C2C2-YABBY | 6 | 1 | 17% |
CAMTA | 5 | 2 | 40% |
EIL | 5 | 2 | 40% |
REM | 5 | 1 | 20% |
DBP | 4 | 1 | 25% |
NLP | 4 | 1 | 25% |
RAV | 4 | 1 | 25% |
RWP-RK | 4 | 2 | 50% |
S1Fa-like | 3 | 1 | 33% |
BES1 | 2 | 1 | 50% |
zf-GRF | 1 | 1 | 100% |
此表数据来源文献,doi: 10.1016/j.cell.2016.04.038。
5.为什么有些基因家族的成功率很低?
有些转录因子需要和其他蛋白形成复合体才能与DNA结合,这些蛋白的风险比较高。
6.一些特殊的样品能不能做,有没有风险?
有两种情况的样品是不能做DAP-seq 实验的,一种情况是没有参考基因组,另一种情况是转录因子不能在体外表达出来,除此之外,我们会做可行性分析报告供您参考。
7.包含重复吗?
包含两个技术重复。
8.做这个蛋白表达的时候,使用的什么表达系统?
优先使用真核表达系统进行蛋白表达,如果真核表达系统不能表达成功的话可以沟通换用原核表达系统。
9.植物组织样本取样的时期部位有什么要求?
植物组织样本取样的时期和部位是您根据自己的研究需求确定,不同组织和时期DNA的修饰不同,可能会影响蛋白和DNA的结合。
10.DAP-seq试验结果的可靠性如何,是否能通过验证试验做出来?
可以参考2019-JXB-Populus euphratica PeWRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance这篇文献,文献中是使用DAP-seq技术,在基因组水平上,鉴定了PeWRKY1转录因子与胡杨基因组DNA的结合位点信息,并通过酵母单杂交、EMSA、荧光素酶检测系统验证了这一结果。
11.DAP-seq跟ChIP-seq有何区别,DAP-seq的优势表现在哪里?
DAP-seq和ChIP-seq的区别:

DAP-seq的优势:不需要针对每个转录因子制备特异性抗体,快速、高通量、节约时间成本。
12、DAP-seq用的input是什么,为什么选这个作为对照呢?
Input对照是用的亲和纯化前的文库,目的是降低背景噪音,我们用的Input和2016年发表在Cell(DAP Seq-Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape)上的论文是一致的。
13、为什么实验中表达的有些蛋白比理论值偏大?
很多蛋白表达出来比理论值大一些,因为有一些翻译后修饰,很多情况都是这样的,原核表达也有这类情况,比如拟南芥SnRK蛋白激酶,预测40 kd,通过原核表达,实际分子量是60 kd。
| 题目 | 期刊 | IF | 发表日期 |
ARF3-mediated auxin signaling is essential for sex determination in cucumber | Science | 45.8 | 2025 |
Accelerated fatty acid biosynthesis by an RCC1-domain protein enables record-high productivity in Schizochytrium | Chem Eng J | 13.2 | 2025 |
An incoherent feed-forward loop coordinates nitrate uptake and tillering in wheat | Mol Plant | 24.1 | 2025 |
GmMYB4 Positively Regulates Isoflavone Biosynthesis via the GmMAPK6-GmMYB4-MBW Module in Soybean | Plant Biotechnol J | 10.5 | 2025 |
An apoplastic fungal effector disrupts N-glycosylation of ZmLecRK1, inducing its degradation to suppress disease resistance in maize | Nat Plants | 13.6 | 2025 |
SlBES1-mediated brassinosteroid signaling suppresses flavonoid biosynthesis in tomato fruit | Plant Commun | 11.6 | 2025 |
Heterologous expression of the barley-specific HvbZIP87 transcription factor in wheat enhances broad-spectrum disease resistance with balanced yield | J Adv Res | 11.4 | 2025 |
| Chromosome-level genome assembly assisting for dissecting mechanism of anthocyanin regulation in kiwifruit (Actinidia arguta) | Mol Hortic | 10 | 2025 |
The Intronic Structure Variation of Rapeseed BnaC3.LEAFY Regulates the Timing of Inflorescence Formation and Flowering | Plant Commun | 9.4 | 2025 |
OsNLP3 and OsPHR2 orchestrate direct and mycorr-hizal pathways for nitrate uptake by regulating NAR2.1-NRT2s complexes in rice | PNAS | 9.4 | 2025 |
Volatilome-based GWAS identifies OsWRKY19 and OsNAC021 as key regulators of rice aroma | Mol Plant | 17.1 | 2024 |
MYB-related transcription factors control chloroplast biogenesis | Cell | 45.5 | 2024 |
Arabidopsis WRKY1 promotes monocarpic senescen -ce by integrative regulation of flowering, leaf sene-scence and nitrogen remobilization | Mol Plant | 17.1 | 2024 |
The MdHSC70-MdWRKY75 module mediates basal apple thermotolerance by regulating the expression of heat shock factor genes | Plant Cell | 11.6 | 2024 |







