- 技术简介
- 实验数据
- 发表文献
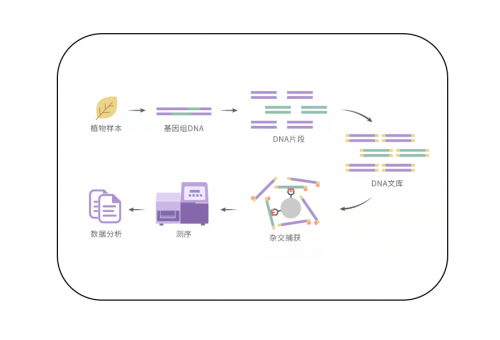
蓝景科信将探针杂交捕获技术和NGS技术结合,提供被子植物353个单拷贝核基因靶向捕获测序服务。为研究被子植物的系统进化和分类、植物适应性机制、种群进化历史、种群的结构、基因渗透和漂移提供高效的整体解决方案。靶向捕获测序技术服务,使用生物素化的探针,捕获被子植物353个单拷贝核基因。您可以提供新鲜植物样本,腊叶标本或者古老的博物馆标本,我们负责DNA提取、文库构建、靶向捕获、测序和数据分析。
技术流程


Fig 1. Heatmap of Gene Recovery Efficiency. Each row is one sample, and each column is one gene. Colors indicate the percentage of the target length (calculated by the mean length of all k-medoid transcripts for each gene) recovered.

Fig 2. Total Length of Sequence Recovery for Both Coding and Non-coding Regions Across 353 Loci for 42 Angiosperm Species. Reads were mapped back to either coding sequence (yellow) or coding sequence plus flanking non-coding (i.e. intron) sequence (purple)… The total length of coding sequence targeted was 260,802 bp. The median recovery of coding sequence was 137,046 bp and the median amount of non-coding sequence recovered was 216,816 bp (with at least 8x depth of coverage).
参考文献:
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GK, Baker WJ, Wickett NJ. A Universal Probe Set for Targeted Sequencing of 353 Nuclear Genes from Any Flowering Plant Designed Using k-Medoids Clustering. Syst Biol. 2019. 68(4):594-606. doi: 10.1093/sysbio/syy086.
使用被子植物353个单拷贝核基因捕获探针进行靶向捕获测序,发表的部分文献:
Antonelli, A., Clarkson, J.J., Kainulainen, K., Maurin, O., Brewer, G.E., Davis, A.P., … Baker, W.J. (2021). Settling a family feud: a high-level phylogenomic framework for the Gentianales based on 353 nuclear genes and partial plastomes. American Journal of Botany 108, 1143–1165.
Baker, W.J., Bailey, P., Barber, V., Barker, A., Bellot, S., Bishop, D., … Forest, F. (2021). A Comprehensive Phylogenomic Platform for Exploring the Angiosperm Tree of Life. Systematic Biology.
Brewer, G.E., Clarkson, J.J., Maurin, O., Zuntini, A.R., Barber, V., Bellot, S., … Baker, W.J. (2019). Factors Affecting Targeted Sequencing of 353 Nuclear Genes From Herbarium Specimens Spanning the Diversity of Angiosperms. Front. Plant Sci. 0.
Buerki, S., Callmander, M.W., Acevedo-Rodriguez, P., Lowry, P.P., Munzinger, J., Bailey, P., … Forest, F. (2021). An updated infra-familial classification of Sapindaceae based on targeted enrichment data. American Journal of Botany 108, 1234–1251.
Clarkson, J.J., Zuntini, A.R., Maurin, O., Downie, S.R., Plunkett, G.M., Nicolas, A.N., … Baker, W.J., (2021). A higher-level nuclear phylogenomic study of the carrot family (Apiaceae). American Journal of Botany 108, 1252–1269.
Gaynor, M.L., Fu, C.-N., Gao, L.-M., Lu, L.-M., Soltis, D.E., Soltis, P.S. (2020). Biogeography and ecological niche evolution in Diapensiaceae inferred from phylogenetic analysis. Journal of Systematics and Evolution 58, 646–662.
Hendriks, K.P., Mandáková, T., Hay, N.M., Ly, E., Huysduynen, A.H. van, Tamrakar, R., … Bailey, C.D. (2021). The best of both worlds: Combining lineage-specific and universal bait sets in target-enrichment hybridization reactions. Applications in Plant Sciences 9.
Johnson, M., Pokorny, L., Dodsworth, S., Botigue, L.R., Cowan, R.S., Devault, A., … Wickett, N. (2019). A Universal Probe Set for Targeted Sequencing of 353 Nuclear Genes from Any Flowering Plant Designed Using k-medoids Clustering. Systematic Biology 68(4): 594-606.
Larridon, I., Villaverde, T., Zuntini, A.R., Pokorny, L., Brewer, G.E., Epitawalage, N., … Baker, W.J. (2020). Tackling Rapid Radiations With Targeted Sequencing. Frontiers in Plant Science 10: 1.
Larridon, I., Zuntini, A.R., Barrett, R.L., Wilson, K.L., Bruhl, J.J., Goetghebeur, P., … Roalson, E.H. (2021). Resolving generic limits in Cyperaceae tribe Abildgaardieae using targeted sequencing. Botanical Journal of the Linnean Society 196, 163–187.
Larridon, I., Zuntini, A.R., Léveillé-Bourret, É., Barrett, R.L., Starr, J.R., Muasya, A.M., … Baker, W.J. (2021). A new classification of Cyperaceae (Poales) supported by phylogenomic data. Journal of Systematics and Evolution 59, 852–895.
Lee, A.K., Gilman, I.S., Srivastav, M., Lerner, A.D., Donoghue, M.J., Clement, W.L. (2021). Reconstructing Dipsacales phylogeny using Angiosperms353: issues and insights. American Journal of Botany 108, 1122–1142.
Maurin, O., Anest, A., Bellot, S., Biffin, E., Brewer, G., Charles-Dominique, T., … Lucas, E. (2021). A nuclear phylogenomic study of the angiosperm order Myrtales, exploring the potential and limitations of the universal Angiosperms353 probe set. American Journal of Botany 108(7): 1087–1111.
Murphy, B., Forest, F., Barraclough, T., Rosindell, J., Bellot, S., Cowan, R., … Cheek, M. (2020). A phylogenomic analysis of Nepenthes (Nepenthaceae). Molecular Phylogenetics and Evolution 144, 106668.
Ogutcen, E., Christe, C., Nishii, K., Salamin, N., Möller, M., Perret, M. (2021). Phylogenomics of Gesneriaceae using targeted capture of nuclear genes. Molecular Phylogenetics and Evolution 157, 107068.
Ottenlips, M.V., Mansfield, D.H., Buerki, S., Feist, M.A.E., Downie, S.R., Dodsworth, … Smith, J.F. (2021). Resolving species boundaries in a recent radiation with the Angiosperms353 probe set: the Lomatium packardiae/L. anomalum clade of the L. triternatum (Apiaceae) complex. American Journal of Botany 108, 1217–1233.
Pérez-Escobar, O.A., Dodsworth, S., Bogarín, D., Bellot, S., Balbuena, J.A., Schley, R.J., … Baker, W.J. (2021). Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. American Journal of Botany 108, 1166–1180.
Pillon, Y., Hopkins, H.C.F., Maurin, O., Epitawalage, N., Bradford, J., Rogers, Z.S., … Forest, F. (2021). Phylogenomics and biogeography of Cunoniaceae (Oxalidales) with complete generic sampling and taxonomic realignments. American Journal of Botany 108, 1181–1200.
Shah, T., Schneider, J.V., Zizka, G., Maurin, O., Baker, W., Forest, F., … Larridon, I., (2021). Joining forces in Ochnaceae phylogenomics: a tale of two targeted sequencing probe kits. American Journal of Botany 108, 1201–1216.
Shee, Z. Q., D. G. Frodin, R. Cámara-Leret, L. Pokorny. (2020). Reconstructing the Complex Evolutionary History of the Papuasian Schefflera Radiation Through Herbariomics. Frontiers in Plant Science 11: 258.
Siniscalchi, C.M., Hidalgo, O., Palazzesi, L., Pellicer, J., Pokorny, L., Maurin, O., … Mandel, J.R. (2021). Lineage-specific vs. universal: A comparison of the Compositae1061 and Angiosperms353 enrichment panels in the sunflower family. Applications in Plant Sciences 9.
Starr, J.R., Jiménez-Mejías, P., Zuntini, A.R., Léveillé-Bourret, É., Semmouri, I., Muasya, M., … Larridon, I. (2021). Targeted sequencing supports morphology and embryo features in resolving the classification of Cyperaceae tribe Fuireneae s.l. Journal of Systematics and Evolution 59, 809–832.
Thomas, A.E., Igea, J., Meudt, H.M., Albach, D.C., Lee, W.G., Tanentzap, A.J. (2021). Using target sequence capture to improve the phylogenetic resolution of a rapid radiation in New Zealand Veronica. American Journal of Botany 108, 1289–1306.
Thomas, S.K., Liu, X., Du, Z.-Y., Dong, Y., Cummings, A., Pokorny, L., … Leebens-Mack, J.H. (2021). Comprehending Cornales: phylogenetic reconstruction of the order using the Angiosperms353 probe set. American Journal of Botany 108, 1112–1121.
Ufimov, R., Zeisek, V., Píšová, S., Baker, W.J., Fér, T., Loo, M., … Schmickl, R. (2021). Relative performance of customized and universal probe sets in target enrichment: A case study in subtribe Malinae. Applications in Plant Sciences 9, e11442.
Van Andel, T., Veltman, M. A., Bertin, A., Maat, H., Polime, T., Hille Ris Lambers, D., … Manzanilla, V.. (2019). Hidden Rice Diversity in the Guianas. Frontiers in Plant Science 10: 1161.
Wenzell, K.E., McDonnell, A.J., Wickett, N.J., Fant, J.B., Skogen, K.A. (2021). Incomplete reproductive isolation and low genetic differentiation despite floral divergence across varying geographic scales in Castilleja. American Journal of Botany 108, 1270–1288.
Zuntini, A.R., Frankel, L.P., Pokorny, L., Forest, F., Baker, W.J. (2021). A comprehensive phylogenomic study of the monocot order Commelinales, with a new classification of Commelinaceae. American Journal of Botany 108, 1066–1086.







